Pluripotent Cell Technology services portfolio
Gene targeting in mESC
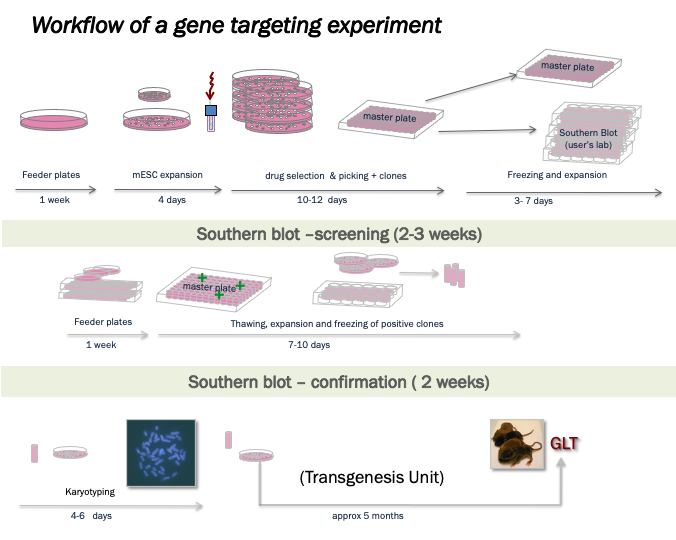
Currently, the Pluripotent Cell Technology Unit continues to offer basic support in the generation of genetically modified mice via homologous recombination. With the purpose of accelerating and facilitating genetic modification projects, the Unit offers to users a wide range of services:
- Advice on the targeting vector design and screening strategy
- Electroporation in mESC of the targeting vector
- Screening by Southern blot or PCR
- Expansion of the genetically modified clones and verification by Southern blot of the recombination events
- Karyotype analysis of modified clones until their delivery to the Transgenesis Unit for aggregation or microinjection sessions
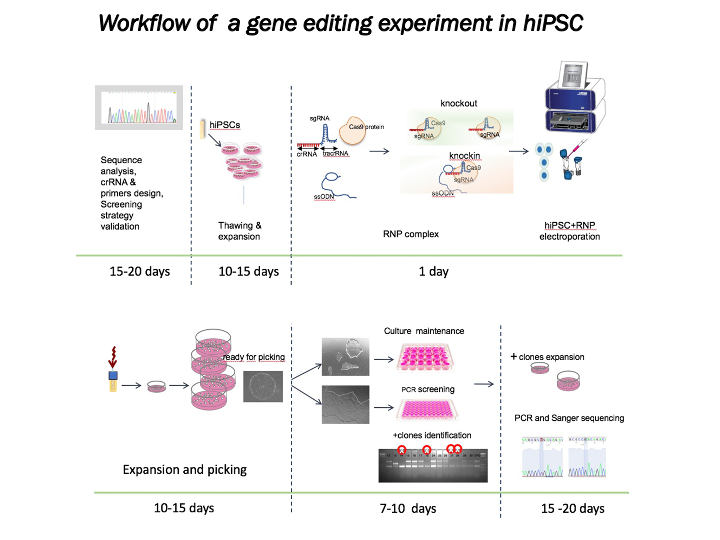
Gene editing in mESC and hiPSC trough CRISPR/Cas9 System
We offer advice to the investigators for optimal CRISPR/Cas9 design and screening strategy:
- Consultation and validation of crRNA
- Electroporation of CRISPR/Cas9 plasmids or ribonucleoprotein complexes (RNP)
- Screening by PCR to identify modified clones
- Sanger sequencing analysis to confirm mutant clones
- Expansion and banking of the genetically modified clones
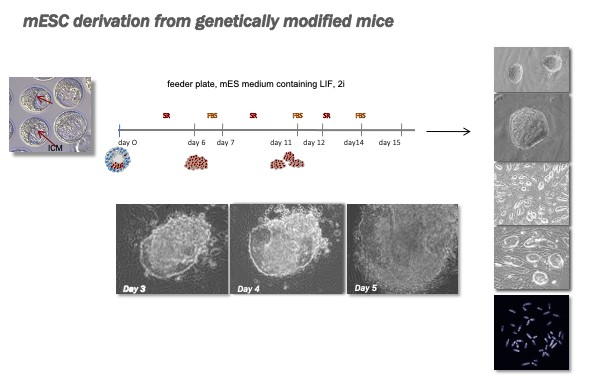
mESC derivation and differentiation
The PCTUnit staff derive mESC from blastocysts isolated from mutant mice and provide support to researchers in carrying out differentiation protocols to cardiac lineage.
Hatching and expansion of the ICM maintained onto feeder layers of cells and in appropriate media.
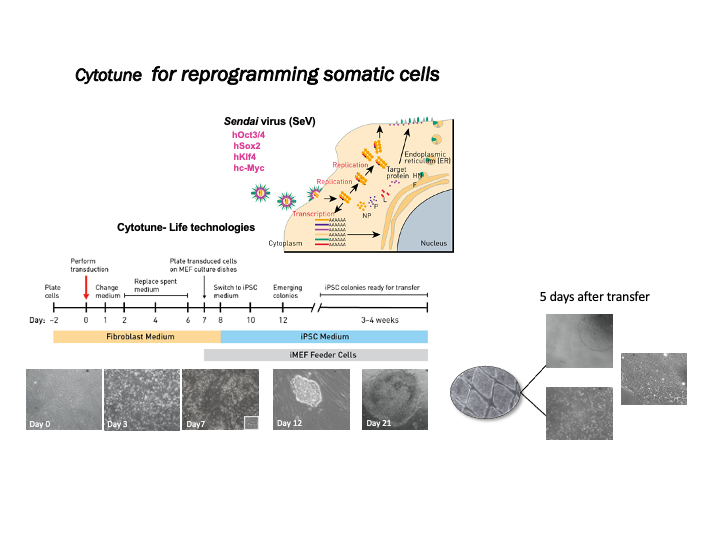
Reprogramming somatic cells to hiPSC
The PCTUnit staff offer essential support in the generation of cardiovascular disease models by establishing hiPSC lines derived from dermal fibroblasts using the non-integrative system of Sendai virus (Cytotune v2). Applying the CRISPR/Cas9 gene editing system we facilitate the creation of isogenic lines. The in vitro comparative analysis between wildtype and mutant cells enable to isolate the effect of the mutation.
Note: Before being used in reprogramming experiments, it is mandatory that all patient cell lines comply with the regulatory requirements indicated by the ISCIII Ethics Committee.
The specific activities of the hiPSC facility include:
- hiPSC reprogramming from somatic cells using non-integrative Sendai virus system.
- Performing quality-control analysis of hiPSC cell lines:
- Testing for the absence of SeV by RT-PCR
- Alkaline phosphatase assay, immunostaining for pluripotency markers (Octa4, Sox2, Nanog, etc.)
- Embryoid body formation and immunostaining for differentiation markers of the three cell lineages
Human Induced Pluripotent Stem Cell Derived cardiomyocytes (hiPSC-CM)
Setting up differentiation protocols from hiPSC is expensive and time-consuming. Standardization of culture conditions and availability of validated reagents will inevitably entail an optimization of time and economic resources. Taking into account the numerous methodologies available in the field of cardiac differentiation and the possibility of specific subtype enrichment, each differentiation project will be discussed with the investigators in order to achieve the required objective.
As a general rule, monolayers differentiation program of hiPSC-CM derived is based on the following steps:
- Expansion of the hiPSC clone
- Optimization of cell density and reagents concentrations
- Cardiomyocytes purifications using MACS-technology fine-tuned by Miltenyi Biotec.
- Maturation on silicone membrane (PDMS) as described in Rocha AM et al. Sci Rep 7: 13834.







