Cell: CNIC Scientists develop new methods for analyzing gene function
The study, published in Cell, will allow any researcher to induce multispectral genetic mosaics in vertebrate research models such as mice and zebrafish
Scientists at the Centro Nacional de Investigaciones Cardiovasculares Carlos III (CNIC) have developed new methods to produce and analyze genetic mosaics. In these mosaics, tissues contain various groups of cells with different known genotypes, permitting study of the differences that these genotypes generate in cell behavior. The new methods, published in Cell, will allow any researcher to induce multispectral genetic mosaics in vertebrate models such as mice and zebrafish. The technology will help advance understanding of the function and interaction of genes, at high spatio-temporal resolution, during development and disease. Genes encode the information needed to synthesize proteins, the functional building blocks of our cells. Improved methods for studying gene function will allow researchers to “increase knowledge about how the genome operates in the multiple cell types that make up our bodies and understand gene interaction networks and their regulatory hierarchies.” This knowledge, moreover, is crucial for the design of efficient therapeutic strategies to modify or correct genetic activity in disease.
The ability to modify gene activity has radically changed approaches to the study of biological processes. Today, most researchers analyze the function of one gene at a time by modifying its activity—increasing it or eliminating it—in all or a subset of cells of an organ or animal. With this approach, the understanding of gene function is obtained through the analysis of the changes that a single genetic modification produces in the development, function, or disease of the organ or animal.
As lead author Rui Benedito explained, in experimental approaches using inducible genetic mosaics, the induced genetic changes take place only in some cells of the organism (mutant cells), while the other cells remain unaltered (normal cells). “The use of inducible genetic mosaics is very important because it enables us to study how cells with different genotypes behave in the same environment, so that any differences can be attributed to the induced genetic alteration.” This approach to the study of gene function is often more precise and informative than the use of classical genetics, in which modification of the target gene in all cells of the organism can generate secondary alterations that cannot be controlled temporally or spatially and that could distort interpretation of the function of the gene in the process under study.
This new technology will provide information at high temporal and cellular resolution about gene functions and interactions during organ development and in disease
Genetic mosaics have been used extensively in the fruit fly due to the ease of performing mitotic recombination in this organism, and have revolutionized the study of gene function and cell biology. However, it has so far been technically much more challenging to induce and analyze genetic mosaics in the mouse, the most widely used model organism in biomedical research.
To generate genetic mosaics in mice, scientists in the Molecular Genetics of Angiogenesis lab at the CNIC have developed new molecular biology and transgenesis methods. These methods allow the simultaneous induction, in a single mouse, of multiple mosaic genetic modifications associated with the expression of distinct fluorescent markers that can be detected by high-resolution multispectral microscopy.
Information derived from this technology could be important to designing efficient therapeutic strategies to modify or correct genetic activity in disease
To make these new genetic tools usable by other research groups, the CNIC team first developed an open-source DNA engineering strategy that greatly simplifies the production of large and complex DNA constructs containing multiple target genes and fluorescent marker genes in frame. The team also developed new CRISPR/Cas9 methods to introduce these DNA molecules into embryonic stem cells or zygotes, simplifying the rapid and reliable generation of transgenic mouse mosaics.
With these new genetic tools and transgenic mice, the CNIC team was able to study the function of multiple genes in the same tissue by simultaneous fluorescence microscopy of up to 15 color-coded cell populations, each expressing a unique combination of genes. According to Rui Benedito, “this technology for the first time allows combinations of multiple genetic mosaics to be induced and analyzed in different cells of the same mouse. Because the different cell populations are induced in the same tissue, and can be imaged simultaneously, an analysis of how their behaviors differ can provide precise data and new insights into the function of the genes under study. This approach has real advantages over established procedures for genetic analysis, which require comparison of cells present in different tissues or environments, since the analysis must be done with independent animals that have or do not have a given genetic modification.”
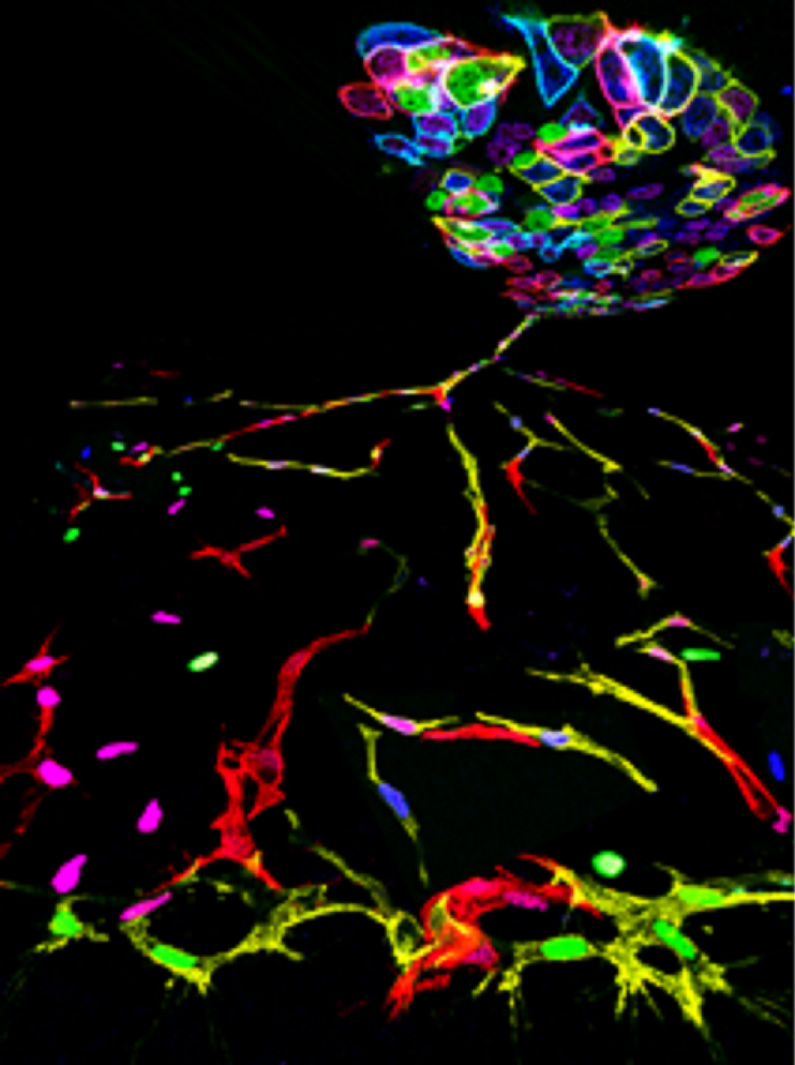
Figure 1: Pontes-Quero et al. have developed new geneticools, cell lines, and transgenic mice that can be used to study the function of genes with a high level of retrospective clonal and phenotypic resolution. The image shows an artistic representation based on real microscopy images, illustrating an induced fluorescent-genetic mosaic in a cluster of embryonic stem cells from which endothelial cells differentiate, proliferate, and sprout to form new blood vessels. Individual cells differ in their proliferative and migratory capacity.
Pontes-Quero, S., Heredia, L., Casquero-Garcia, V., Fernandez-Chacon, M., Luo, W., Hermoso, A., . . . Benedito, R. (2017). Dual ifgMosaic: A Versatile Method for Multispectral and Combinatorial Mosaic Gene-Function Analysis. Cell, 170(4), 800-814 e818. doi:10.1016/j.cell.2017.07.031











